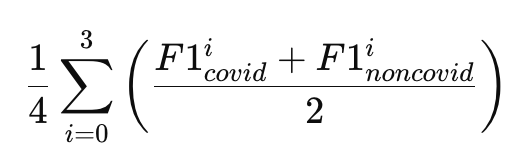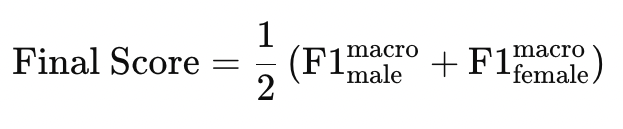If you use the above data, you must cite all following papers and the white paper that will be distributed at a later stage:
D. Kollias, et. al.: “Domain adaptation, Explainability & Fairness in AI for Medical Image Analysis: Diagnosis of COVID-19 based on 3-D Chest CT-scans“, CVPR 2024
@inproceedings{kollias2024domain,title={Domain adaptation explainability \& fairness in ai for medical image analysis: Diagnosis of covid-19 based on 3-d chest ct-scans},author={Kollias, Dimitrios and Arsenos, Anastasios and Kollias, Stefanos},booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},pages={4907--4914},year={2024}}
D. Kollias, et. al.: “Sam2clip2sam: Vision language model for segmentation of 3d ct scans for covid-19 detection“, EAI ETPHT 2024
@article{kollias2024sam2clip2sam,title={Sam2clip2sam: Vision language model for segmentation of 3d ct scans for covid-19 detection},author={Kollias, Dimitrios and Arsenos, Anastasios and Wingate, James and Kollias, Stefanos},journal={arXiv preprint arXiv:2407.15728},year={2024}}
D. Gerogiannis, et. al.: “Covid-19 computer-aided diagnosis through ai-assisted ct imaging analysis: Deploying a medical ai system“, IEEE ISBI 2024
@inproceedings{gerogiannis2024covid,title={Covid-19 computer-aided diagnosis through ai-assisted ct imaging analysis: Deploying a medical ai system},author={Gerogiannis, Demetris and Arsenos, Anastasios and Kollias, Dimitrios and Nikitopoulos, Dimitris and Kollias, Stefanos},booktitle={2024 IEEE International Symposium on Biomedical Imaging (ISBI)},pages={1--4},year={2024},organization={IEEE}}
D. Kollias, et. al.: “AI-Enabled Analysis of 3-D CT Scans for Diagnosis of COVID-19 & its Severity“, IEEE ICASSP 2023
@inproceedings{kollias2023ai, title={AI-Enabled Analysis of 3-D CT Scans for Diagnosis of COVID-19 \& its Severity}, author={Kollias, Dimitrios and Arsenos, Anastasios and Kollias, Stefanos}, booktitle={2023 IEEE International Conference on Acoustics, Speech, and Signal Processing Workshops (ICASSPW)}, pages={1–5}, year={2023}, organization={IEEE}}
A. Arsenos, et. al.: “Data-Driven Covid-19 Detection Through Medical Imaging“, IEEE ICASSP 2023
@inproceedings{arsenos2023data, title={Data-Driven Covid-19 Detection Through Medical Imaging}, author={Arsenos, Anastasios and Davidhi, Andjoli and Kollias, Dimitrios and Prassopoulos, Panos and Kollias, Stefanos}, booktitle={2023 IEEE International Conference on Acoustics, Speech, and Signal Processing Workshops (ICASSPW)}, pages={1–5}, year={2023}, organization={IEEE}}
D. Kollias, et. al.: “A deep neural architecture for harmonizing 3-D input data analysis and decision making in medical imaging“, Neurocomputing 2023
@article{kollias2023deep,title={A deep neural architecture for harmonizing 3-D input data analysis and decision making in medical imaging}, author={Kollias, Dimitrios and Arsenos, Anastasios and Kollias, Stefanos}, journal={Neurocomputing}, volume={542}, pages={126244}, year={2023}, publisher={Elsevier}}
D. Kollias, et. al.: “AI-MIA: COVID-19 Detection & Severity Analysis through Medical Imaging“, ECCV 2022
@inproceedings{kollias2022ai, title={Ai-mia: Covid-19 detection and severity analysis through medical imaging}, author={Kollias, Dimitrios and Arsenos, Anastasios and Kollias, Stefanos}, booktitle={European Conference on Computer Vision}, pages={677–690}, year={2022}, organization={Springer}}
A. Arsenos, et. al.: “A Large Imaging Database and Novel Deep Neural Architecture for Covid-19 Diagnosis“, IVMSP 2022
@inproceedings{arsenos2022large, title={A Large Imaging Database and Novel Deep Neural Architecture for Covid-19 Diagnosis}, author={Arsenos, Anastasios and Kollias, Dimitrios and Kollias, Stefanos}, booktitle={2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP)}, pages={1–5}, year={2022}, organization={IEEE} }
D. Kollias, et. al.: “MIA-COV19D: COVID-19 Detection through 3-D Chest CT Image Analysis“, ICCV 2021
@inproceedings{kollias2021mia, title={Mia-cov19d: Covid-19 detection through 3-d chest ct image analysis}, author={Kollias, Dimitrios and Arsenos, Anastasios and Soukissian, Levon and Kollias, Stefanos}, booktitle={Proceedings of the IEEE/CVF International Conference on Computer Vision}, pages={537–544}, year={2021} }
D. Kollias, et. al.: “Deep transparent prediction through latent representation analysis”, 2020
@article{kollias2020deep, title={Deep transparent prediction through latent representation analysis}, author={Kollias, Dimitrios and Bouas, N and Vlaxos, Y and Brillakis, V and Seferis, M and Kollia, Ilianna and Sukissian, Levon and Wingate, James and Kollias, S}, journal={arXiv preprint arXiv:2009.07044}, year={2020}}
D. Kollias, et. al.: “Transparent Adaptation in Deep Medical Image Diagnosis”, TAILOR 2020
@inproceedings{kollias2020transparent, title={Transparent Adaptation in Deep Medical Image Diagnosis.}, author={Kollias, Dimitris and Vlaxos, Y and Seferis, M and Kollia, Ilianna and Sukissian, Levon and Wingate, James and Kollias, Stefanos D}, booktitle={TAILOR}, pages={251–267}, year={2020}}
D. Kollias, et. al.: “Deep neural architectures for prediction in healthcare”, CIS 2018
@article{kollias2018deep, title={Deep neural architectures for prediction in healthcare}, author={Kollias, Dimitrios and Tagaris, Athanasios and Stafylopatis, Andreas and Kollias, Stefanos and Tagaris, Georgios}, journal={Complex \& Intelligent Systems}, volume={4}, number={2}, pages={119–131}, year={2018}, publisher={Springer}}
Sponsors
The PHAROS-AFE-AIMI Workshop has been generously supported by:
GRNET – National Infrastructures for Research and Technology

Queen Mary University of London

Digital Environment Research Institute

Institute of Communication and Computer Systems